Assessing Host-Microbiota Relationships
- 9 mars 2018
- 10 min de lecture
Microbes have long had the knack of making themselves at home. In fact, these versatile and adaptable life forms are known to have preceded us by billions of years, exploiting virtually every niche on the planet. Microbes will in all likelihood survive us, too. In the meantime, they are happy to treat us as yet another niche, albeit a fairly complicated one.
Our microbes and our bodies often practice a form of mutual accommodation. It’s a delicate balancing act, one that is essential to our health and well-being. Such an important relationship can hardly be left to chance. It must be understood and, if necessary, managed.
Before we can even think of tipping our microbiome scales—pharmacologically, probiotically, or otherwise—we need to become adept at interrogating the identity and the functionality of our microbial inhabitants. Fortunately, microbiome studies are yielding new insights and leading to the development of novel probes. For example, since it has become clear that the human microbiome contributes more protein-encoding genes than the human genome, scientists have been adapting proteomic techniques to advance microbiome research.
“One can think of the microbiome as an incredibly complex organ in our body,” says Dennis W. Wolan, Ph.D., associate professor of molecular and experimental medicine at The Scripps Research Institute. “It has lots of functionalities and interactions that dictate our health.”
A wealth of phylogenetic analyses helped characterize the microbial ecosystem in various environments on a species level. However, comparatively, understanding the functions that specific microorganisms perform in the niches they colonize has been lagging.
“The real challenge is to understand what they do, what they make, and how they interact with us,” insists Dr. Wolan. An indispensable part of the efforts needed to interrogate these interactions requires an in-depth analysis of the proteins, peptides, and small molecules that are generated by these bacteria and the way in which hosts use them or respond to their presence.
A Metaproteomic Gut Check
To capture the dynamics of the proteins that are made by the microbiome, Dr. Wolan and colleagues introduced two new techniques to metaproteomic analysis. “These are not new techniques in themselves, but they have not been previously applied to study the highly complex bacterial samples by metaproteomics approaches,” comments Dr. Wolan.
The first technique is called stable isotope labeling in mammals (SILAM). It was used by Dr. Wolan’s group to quantitate protein abundances across two mouse cohorts. The experimental cohort, which modeled inflammatory bowel disease, was fed a heavy nitrogen isotope; the control cohort received the normally abundant isotope.
This work marked the first time the incorporation of isotopes from animal feed into the microbiota was studied. “It took only a few days to incorporate the isotope into the microbiota,” notes Dr. Wolan. “This result showed that there is a relatively quick turnover of the microbes in the gut.”
The second technique, activity-based probes, involves the labeling of a protein of interest with a covalent tag, an approach that allows the interacting proteins to be isolated and enriched. “This is important for metaproteomics,” Dr. Wolan explains. “At this time, we are very much limited in the sensitivity of mass spectrometry. As a result, metaproteomic analyses are not digging that deeply into the proteins that are present in samples.”
Although mass spectrometers are becoming more sensitive, they are currently capable of analyzing only about 20% of the most abundant proteins from microbiome samples. To improve metaproteomic study of microbiome samples, Dr. Wolan decided to go beyond the usual approach: tandem mass spectrometry-based shotgun proteomics.
The combination of isotope labeling and shotgun proteomics, Dr. Wolan and colleagues explained, was conceived as a way to deal with the inherent complexity and taxonomic diversity of microbiome samples. The idea was to compare the microbiota from their disease model to the microbiota of an isogenic control, while limiting the genetic and environmental variables that influence microbiome composition. Ultimately, the combination approach helped the investigators interrogate new classes of proteins and find quantitative changes in both host and microbial proteins that could be attributed to the intestinal inflammation.
“We are not finding new phenotypes, neither are we improving the phylogenetic composition,” reports Dr. Wolan. “But what is new with our approach is that we can see more proteins and get a deeper sense of what is in the proteome.”
From over 4,400 protein clusters identified in the microbial proteomic environment, over 270 were differentially expressed between the two animal groups, including proteins of unknown function.
“A lot of the bacteria in the microbiome provide functionalities that interact with the host,” emphasizes Dr. Wolan. “We would like to better understand the composition of enzymes secreted by bacteria and hosts, as well as the crosstalk between the host and the microbes and also among the microbes themselves.”
A Microbiome-Parkinson’s Link
Click Image To Enlarge +
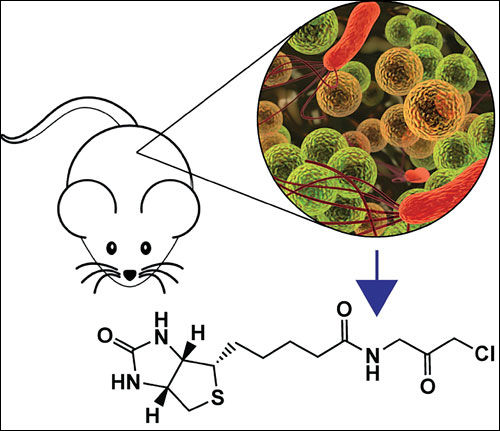
At the California Institute of Technology, Timothy R. Sampson, Ph.D., and other scientists in the laboratory of Sarkis K. Mazmanian, Ph.D., used mice that overexpress a-synuclein to show that gut microbiota are required for the motor deficits, microglia activation, and a-synuclein pathology that are characteristic of Parkinson’s disease.
“Studies that have profiled microbiomes in different conditions are incredibly valuable,” says Sarkis K. Mazmanian, Ph.D., professor of microbiology at California Institute of Technology. “These studies give us insights into microbial community compositions associated with a disease state, a diet, or geography.”
One of the efforts in Dr. Mazmanian’s laboratory is focusing on dissecting the cellular and molecular mechanisms that underlie host-microbiome interactions in physiological and pathological conditions. Animal studies are particularly powerfully positioned to provide insight into the mechanistic basis of associations identified in human clinical studies. Animal and human studies, comments Dr. Mazmanian, are “complementary to each other.”
In one animal study, Dr. Mazmanian and colleagues used mice that overexpressed α-synuclein and displayed symptoms of Parkinson’s disease. The scientists showed that in this model, the gut microbiota regulated motor deficits and neuroinflammation.
Germ-free animals, or animals in which the microbiota was depleted with antibiotics, showed reduced neuronal inclusions and decreased microglial activation and motor deficits. When α-synuclein-overexpressing mice were colonized with microbiota from Parkinson’s disease patients, their physical impairment increased as compared to animals that had been transplanted microbiota from healthy humans.
These findings showed that α-synuclein and dysbiosis, a genetic and an environmental factor, converge in shaping the clinical manifestations of the disease. Administration of microbially produced short-chain fatty acids restored microglial activation and motor dysfunction, providing insight into potential molecular mechanisms of the gut-brain axis dysregulation in this medical condition.
Although the first human diseases linked to perturbations in the microbiota were mostly gastrointestinal and immune-mediated conditions, the spectrum of disorders potentially impacted by the microbiome has expanded over the years and now includes conditions that decades ago were thought to have little in common with dysbiosis. Neurological diseases are one of the most recent conditions to have joined this group.
“Over the next few years, we will likely see an increase in studies linking gut microbes to neurological conditions,” predicts Dr. Mazmanian. Advances in understanding the links between dysbiosis and neurological conditions are fueled by a growing interest and increasing availability and accessibility of molecular techniques, and by a better understanding of the ways in which microbes impact the gut-brain axis.
“One thing that would help research in this area is the development of analytical tools and software interfaces,” notes Dr. Mazmanian. “They would make microbiome data more accessible to people without expertise in bioinformatics, including researchers and clinicians.”
Pathogenic Strains, Worrisome Genes
“The direction that we are pursuing in the next few years is to go from correlations to identifying cause-and-effect relationships,” says Gregory A. Buck, Ph.D., professor of microbiology and immunology at Virginia Commonwealth University School of Medicine. “Once we do that, we can identify and examine predictive biomarkers.”
Many studies have interrogated the microbiome, and they have generated vast amounts of data. To date, however, most of this work has been limited to finding correlations. It has not been very helpful in establishing cause-and-effect relationships. Still, correlations provide a valuable and indispensable starting point to design subsequent experiments powered to examine cause-and-effect relationships. “One can isolate a bacterium that is correlated with an adverse outcome, look at its genome, and find genes that are indicative of adverse outcomes,” explains Dr. Buck.
In a recent study, Dr. Buck and colleagues examined the genomic variation among Mycoplasma hominis strains to identify the genetic elements responsible for the pathogen’s ability to colonize and invade the amniotic cavity and the placenta. Microbial invasion of the amniotic cavity has been associated with spontaneous preterm labor and adverse pregnancy outcomes, and M. hominis, a component of the vaginal microbiota that is sometimes implicated in bacterial vaginosis, has often been isolated from infected fetal membranes and the amniotic fluid.
Dr. Buck and colleagues sequenced the entire genomes of two amniotic fluid isolates and a placental isolate of M. hominis from pregnancies that resulted in preterm births and compared them with the previously sequenced genome of a strain isolated from a healthy individual. This work led to the identification, in the pathogenic strains, of two genes encoding surface membrane proteins. These proteins were truncated in the nonpathogenic strain.
Additionally, the presence of a new gene of unknown function in the pathogenic isolates was significantly associated with the bacterial burden in the amniotic fluid and with preterm birth. “If we know that a particular bacterium with a particular genetic profile in a particular environmental niche might be correlated with disease, we might be able to detect that early,” speculates Dr. Buck. “That is something that could help develop treatments that would impact that particular strain of bacterium or combinations of bacteria.”
One of the prerequisites for performing these comparative studies is the availability of reference genomes. “There are still a lot of reference genomes that are unavailable,” complains Dr. Buck. “Even some of the ones that are available are not included in some of the databases.”
In a more recent prospective study, Dr. Buck and colleagues revealed for the first time that in premature infants, skin-to-skin exposure is associated with a decreased prevalence of Neisseria, Pseudomonas, and Staphylococcus in the saliva, and a more rapid development of the oral microbiota maturity. “A big challenge in a lot of our work and probably other people’s work is that frequently there is a large contamination of bacterial DNA with human DNA,” informs Dr. Buck. “If there was a convenient way to screen out the human DNA before dedicating extensive efforts to sequence analysis, this would help us in many ways.”
Host-Environment Microbe Exchange
“Our study enabled us to confirm previous work that we performed in humans, showing that there is an exchange of bacteria between animals and the environment,” says Jack A. Gilbert, Ph.D., professor of surgery at the University of Chicago and group leader in microbial ecology at the Argonne National Laboratory.
In a study that for the first time examined the skin and fecal microbiota of captive Komodo dragons from several zoos, Dr. Gilbert and colleagues used 16S rRNA sequencing to examine the animals’ salivary, skin, and fecal microbiota along with environmental microbiota. “We focused on trying to understand the microbial interaction that an animal has with the environment,” notes Dr. Gilbert. “The findings we reported could be used to shape future animal care regimens.”
The scientists collected data suggesting that living in closed environments is associated with extensive host-environment microbial sharing. Moreover, the sharing appears to be “circular in nature.” Captive dragons contributed microbes to their environment, and they reacquired the same microbes from their environment, repeating this exchange in an ongoing cycle without other external sources of microbial diversity.
The sharing pattern seen in enclosed environments may be a radical departure from the host-microbial dynamics that prevail under more ecologically natural conditions. In captivity, animals will not receive significant exposure to microbes not already in their enclosure, with unknown consequences for their health. Similar uncertainties may characterize human host-microbial dynamics in domestic environments.
“Technologically the most challenging technical aspect of these studies is trying to figure out where the bacteria came from in the first place in most of the environments we work in, because we don’t start with a clean slate,” informs Dr. Gilbert. Because virtually every ecosystem on the planet is populated with bacteria, studying the dynamics of microbiomes is challenging in terms of finding a control environment.
In one of the studies, Dr. Gilbert and colleagues addressed this limitation by comparatively examining environments before and after they were suddenly reallocated for different purposes. “By looking at environments that change operationally in terms of their human or animal occupancy, we can examine how the new purpose affects the exchange of bacteria between the animal and the environment,” explains Dr. Gilbert.
Manipulating the Microbiota
Click Image To Enlarge +
The laboratory of Ami S. Bhatt, M.D., Ph.D., at Stanford University is focusing on novel genetic and genomic technologies to reveal how microbial genes and gene products impact patient outcomes, especially in the setting of noncommunicable diseases. This image shows a k-mer-based assembly graph from shotgun sequencing data obtained from the stool of a patient with graft-versus-host disease.
“Our laboratory is focusing on novel genetic and genomic technologies to try to better understand how microbial genes and gene products impact patient outcomes, especially in the setting of noncommunicable diseases,” says Ami S. Bhatt, M.D., Ph.D., assistant professor of medicine and genetics at Stanford University School of Medicine. The fundamental hypothesis that drives research efforts in Dr. Bhatt’s group proposes that alterations in the microbiota may impact disease phenotype in several of these conditions, which include cardiometabolic disorders, cancer, and kidney disease.
“That is not to say that the microbiota will cause these diseases in most cases,” notes Dr. Bhatt. “But we believe that we can potentially fine-tune the clinical severity of the disease by manipulating the microbiota.”
Many studies have investigated microbial species that colonize various environmental niches, many of which established correlations to disease. Taking a step forward in dissecting the crosstalk between microbiomes and their hosts, Dr. Bhatt’s group is specifically focusing on the mechanisms that these microorganisms use to impact disease outcome.
“In order to do that, we have to understand which microbes, microbial genes, or gene products are responsible for modifying host biology,” explains Dr. Bhatt. “We use several genomic tools to try and tease that apart.”
Historically, substantial efforts have focused on identifying mutations in the human genome that are associated with disease. However, even if they are identified, these mutations are currently very difficult to manipulate therapeutically.
“In comparison, the microbiome is a potentially modifiable biomarker of disease,” emphasizes Dr. Bhatt. “We have tools that we can develop and use to change that microbiota.” There are several examples that illustrate the possibility of changing the composition of microbial populations, either with beneficial or detrimental effects. Some examples are antimicrobial use, fecal microbiota transplantation, and prebiotic or probiotic therapies.
Critical for developing these interventions is a better understanding of the organization and dynamics of microbial populations. “We need to have higher throughput and more easily applied imaging and visualization approaches,” insists Dr. Bhatt. “They would help us understand the spatial organization of microorganisms within microbial niches.”
Additionally, the vast percentage of the known microbial species cannot currently be cultured in the laboratory, and finding new methods to improve the ability to culture and study these organisms is an important step toward better characterizing microbial populations.
“Single-cell methods as well as long-read and linked-read sequencing technologies may allow us to understand not only who is there in terms of microorganisms, but also get a better handle on which genes are present within which genomes,” suggests Dr. Bhatt.























Commentaires